Binary file parsing in bioinformatics
23 Dec 2016Sometimes I like to get into the weeds of a problem that is only tangentially related to the actual problem I am trying to solve. For example, the other day I was trying to write a smallapplication to serve up BAM alignments to a user based on a location range query. The goal of the project would be to have a server/client that conforms to the draft GA4GH Streaming API specification.
The goals I set out for myself where:
- Serverless environment, which means using Amazon API Gateway, AWS Lambda (Python runtime), and Amazon DynamoDB for the service components, and Amazon S3 for data storage
- Authenticated access to BAM files on S3
- Limit the number requests as much as possible
The tangential rabbit hole I fell into was in developing routines to parse out the data within a BAM index file into DynamoDB for easy lookup. For easier reference, below is the description of the index format pulled from the SAM format specification.
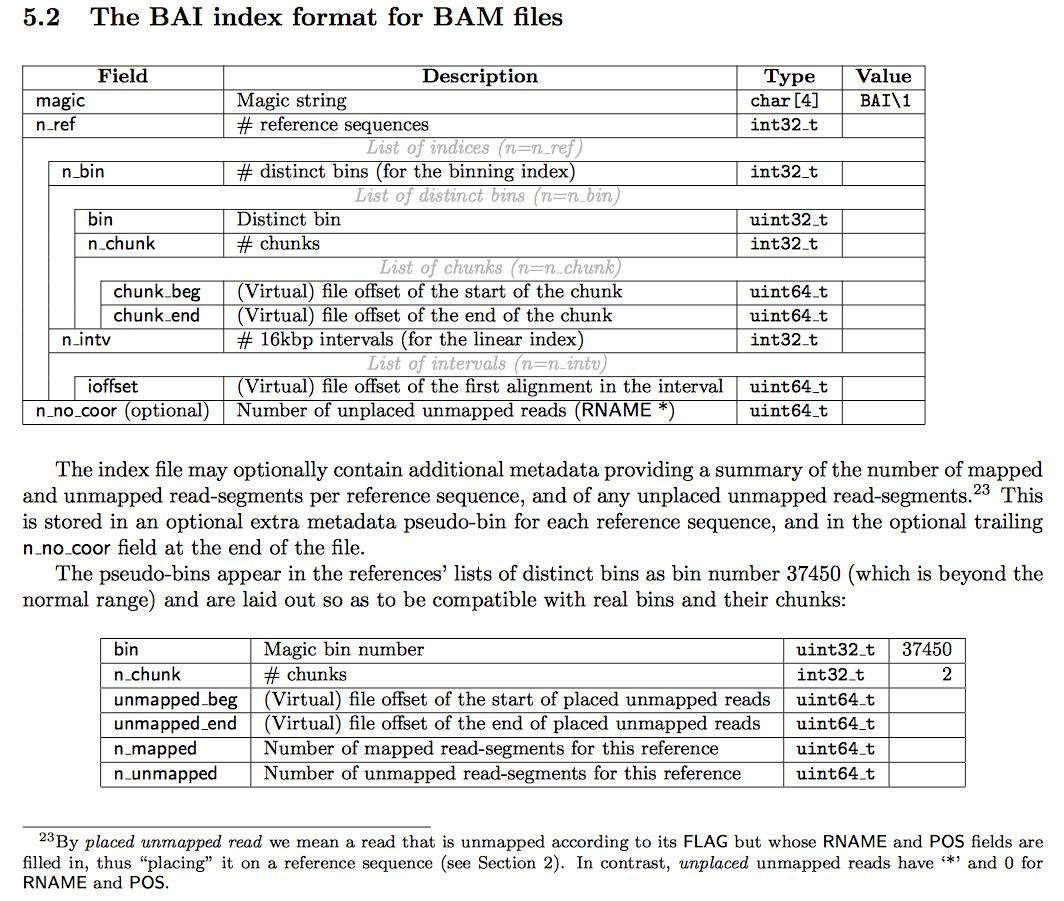
There are no tools that independently inspect this binary format and spit out the information into text, only tools (e.g. PICARD, samtools) that use the index for random access into BAM files. If I wanted that information I would either need to calculate the file offsets myself (as is done in the excellent htsnexus) or implement my own binary file parser for BAI files.
Since I was targeting the Python runtime for Lambda I found two packages that would fit the bill to define the grammer of the binary data into objects that I could easily use, Construct and Kaitai Struct.
Yes, I could have used the core struct Python library, but I wanted a slightly higher level interface to the binary grammer. Both construct and kaitai-struct result in more readable and maintainable code, and both provide concepts for dealing with common data structures that I would have had to deal with on my own. Let’s compare these methods on the binary file format for BAM index files that
Construct
Construct is “a powerful declerative parser (and builder) for binary data”. The library provides primitives for common atomic constructs (e.g. different size integers) and more advanced data structures to allow for defining composite structures. You can read more about it at their site, but I think that an example is best. Let take a look at a chunk which is a pair of virtual file offsets in BAI files. A virtual file offset (section 4.1.1 of the SAM specification) is a tuple of byte offset locations in a BAM file. The first element is the location of the start of a compressed block within a BAM file (coffset), and the second element is the offset of an alignment within the uncompressed data in that block (uoffset). Logically you encode this tuple in a 64 bit integer like so:
# left shift coffset by 16 bits, then OR the uoffset location for the final voffset value
coffset << 16 | uoffset
So a chunk defined using Construct would be:
SVoffset = Struct(
"orig" / Int64ul,
"coffset" / Computed(this.orig >> 16),
"uoffset" / Computed(this.orig ^ (this.coffset << 16))
)
SChunk = Struct(
"chunk_beg" / SVOffset,
"chunk_end" / SVOffset
)
For SVoffset we first need to consume the 64-bit integer from the binary stream, then compute the actual tuple of byte offsets from the original value of the current object using this. A SChunk object is composed of two of the SVoffset objects. For structs that have a variable number of sub-structures, you can also compute the value of the number of elements at runtime using these reference semantics. For example, from the figure above, we see that a bin defines the number of chunks it has by the n_chunk attribute. Thus we can define an Array of type SChunk in the SBin object like so:
SBin = Struct(
"bin" / Int32ul,
"n_chunk" / Int32sl,
"chunks" / Array(this.n_chunk, SChunk)
)
A full example:
import sys
from construct import *
SVoffset = Struct(
"orig" / Int64ul,
"coffset" / Computed(this.orig >> 16),
"uoffset" / Computed(this.orig ^ (this.coffset << 16))
)
SChunk = Struct(
"chunk_beg" / SVoffset,
"chunk_end" / SVoffset
)
SIntv = Struct(
"ioffset" / Int64ul
)
SBin = Struct(
"bin" / Int32ul,
"n_chunk" / Int32sl,
"chunks" / Array(this.n_chunk, SChunk)
)
SRef = Struct(
"n_bin" / Int32sl,
"bins" / Array(this.n_bin, SBin),
"n_intv" / Int32sl,
"intvs" / Array(this.n_intv, SIntv)
)
SBai = Struct(
"magic" / String(4),
"n_ref" / Int32sl,
"refs" / Array(this.n_ref, SRef),
"n_no_coor" / Int64ul
)
## local
io = open(sys.argv[1])
bai = SBai.parse_stream(io)
for r in bai.refs:
for b in r.bins:
c_b = b.chunks[0].chunk_beg
c_e = b.chunks[0].chunk_end
print "bin={0},chunk=[{1},{2}]".format(b.bin, c_b,c_e)
This program, when run against a small test BAI file, produces the following output:
$ python test-construct.py test.bam.bai
bin=4681,chunk=[Container:
orig = 63504384
coffset = 969
uoffset = 63504384,Container:
orig = 1551040512
coffset = 23667
uoffset = 1551040512]
bin=37450,chunk=[Container:
orig = 63504384
coffset = 969
uoffset = 63504384,Container:
orig = 1551040512
coffset = 23667
uoffset = 1551040512]
The result is pretty nice, but I found that on even medium files, the memory explodes as all of the data are read into memory. For example on a 6MB BAI file for NA12878 exome from genome in a bottle, the memory usage was about 475MB and took about 25 seconds on my iMac. AWS Lambda has a ceiling of 1.5GB of memory and for larger BAI we may run into that limit. It also meters by the 100 microseconds, so we want to minimize both of these while we can. While I could have looked into Construct’s lazy parsing features, I decided (e.g. task avoidance level 11) to take a look at another toolchain, Kaitai Struct
Kaitai Struct
In contrast to Construct, Kaitai Struct utilizes a YAML-based grammer file to define the underlying binary structure, and comes with a compiler (written in Scala) to generate files for different languages, including C++, C#, Java, Javascript, Perl, PHP, Ruby, and of course Python. There are also ongoing work to support Rust and Swift.
Let’s take a look at the YAML definition for virtual file offsets and chunks and the resulting Python code.
s_voffset:
seq:
- id: orig
type: u8
instances:
coffset:
value: orig >> 16
uoffset:
value: orig ^ (coffset << 16)
s_chunk:
seq:
- id: chunk_beg
type: voffset
size: 8
- id: chunk_end
type: voffset
size: 8
class SVoffset(KaitaiStruct):
def __init__(self, _io, _parent=None, _root=None):
self._io = _io
self._parent = _parent
self._root = _root if _root else self
self.orig = self._io.read_u8le()
@property
def coffset(self):
if hasattr(self, '_m_coffset'):
return self._m_coffset
self._m_coffset = (self.orig >> 16)
return self._m_coffset
@property
def uoffset(self):
if hasattr(self, '_m_uoffset'):
return self._m_uoffset
self._m_uoffset = (self.orig ^ (self.coffset >> 16))
return self._m_uoffset
class SChunk(KaitaiStruct):
def __init__(self, _io, _parent=None, _root=None):
self._io = _io
self._parent = _parent
self._root = _root if _root else self
self._raw_chunk_beg = self._io.read_bytes(8)
io = KaitaiStream(BytesIO(self._raw_chunk_beg))
self.chunk_beg = self._root.Voffset(io, self, self._root)
self._raw_chunk_end = self._io.read_bytes(8)
io = KaitaiStream(BytesIO(self._raw_chunk_end))
self.chunk_end = self._root.Voffset(io, self, self._root)
In both Construct and Kaitai we see that there are specific trigers for when data are consumed from the byte stream, and when values are computed. In the case, of Kaitai, we use their notion of Instances as the means to define computed properties. Instances can do other things, but I limited my use to this aspect.
Also notice that in contrast to Construct, I had to define the size of the stream that a chunk consumed to pass to the voffset parsing routine, and this is reflected in the python code which pre-fetches the bytes for the SVoffset object. An example script looks pretty similar.
import sys
import bai
s = bai.Bai.from_file(sys.argv[1])
for rn,r in enumerate(s.refs):
for bi,b in enumerate(r.bin):
for ci,c in enumerate(b.chunks):
c_b = [c.chunk_beg.coffset, c.chunk_beg.uoffset]
c_e = [c.chunk_end.coffset, c.chunk_end.uoffset]
print "bin={0},chunk=[{1},{2}]".format(b.bin, c_b, c_e)
$ python test-bai-kt.py test.bam.bai
bin=4681,chunk=[[969, 63504384],[23667, 1551040512]]
bin=37450,chunk=[[969, 63504384],[23667, 1551040512]]
bin=37450,chunk=[[0, 223],[0, 48]]
When I ran this script ran on the NA12878 exome BAM index, it took roughly 900Mb at peak, but only 7 seconds of wall time. So much faster than Construct but it hogs way more memory. Kaitai Stuct does not really provide a grammer for stream or lazy processing, but each language runtime does provide access to a buffer system, and the generated code give good example of how to use it (e.g. KaitaiStream) so one could parse a stream as needed to feed into KaitaiStruct as necessary. Also I would assume that using the C++ STL target would reduce the memory overhead that Python strings incur.
Well that was a fun tangent. I end this post with the complete Kaitai Struct YAML grammer for BAI files. For bonus points I also describe BGZF and BAM, but have tested these so use at your own risk. Enjoy!
meta:
id: bai
file-extension: bai
endian: le
seq:
- id: magic
type: str
size: 4
encoding: UTF-8
contents: BAI\1
- id: n_ref
type: s4
- id: refs
type: ref
repeat: expr
repeat-expr: n_ref
- id: n_no_coor
type: u8
types:
voffset:
seq:
- id: orig
type: u8
instances:
coffset:
value: orig >> 16
uoffset:
value: orig ^ (coffset << 16)
chunk:
seq:
- id: chunk_beg
type: voffset
size: 8
- id: chunk_end
type: voffset
size: 8
bin:
seq:
- id: bin
type: u4
- id: n_chunk
type: s4
- id: chunks
type: chunk
repeat: expr
repeat-expr: n_chunk
ref:
seq:
- id: n_bin
type: s4
- id: bins
type: bin
repeat: expr
repeat-expr: n_bin
- id: n_intv
type: s4
- id: ioffset
type: u8
repeat: expr
repeat-expr: n_intv
# Not used currently, bought here for completeness
pseudo_bin:
seq:
- id: bin
type: u4
contents: '37450'
- id: n_chunk
type: s4
contents: '2'
- id: unmapped_beg
type: voffset
size: 8
- id: unmapped_end
type: voffset
size: 8
- id: n_mapped
type: u8
- id: n_unmapped
type: u8
meta:
id: bam
file-extension: bam
endian: le
seq:
- id: header
type: header
- id: refs
type: ref
repeat: expr
repeat-expr: header.n_ref
- id: algns
type: algn
size-eos: true
repeat: eos
types:
header:
seq:
- id: magic
type: str
size: 4
contents: BAM\1
encoding: UTF-8
- id: l_text
type: s4
- id: text
type: str
size: l_text
encoding: UTF-8
- id: n_ref
type: s4
ref:
seq:
- id: l_name
type: s4
- id: name
type: str
size: l_name
encoding: UTF-8
- id: l_ref
type: s4
algn:
seq:
- id: block_size
type: s4
- id: ref_id
type: s4
- id: pos
type: s4
- id: bin_mq_nl
type: u4
- id: flag_nc
type: u4
- id: l_seq
type: s4
- id: next_ref_id
type: s4
- id: next_pos
type: s4
- id: tlen
type: s4
- id: read_name
type: str
size: l_read_name
encoding: UTF-8
- id: cigar
type: u4
repeat: expr
repeat-expr: no_cigar_op
- id: seq
type: u1
repeat: expr
repeat-expr: l_seq + 1 / 2
- id: qual
type: str
size: l_seq
encoding: UTF-8
- id: tags
type: str
size: block-size - (32 * 9) - l_read_name - (32 * no_cigar_op) - (8 * (l_seq + 1 / 2)) - (8 * l_seq)
encoding: UTF-8
instances:
bin:
value: bin_mq_nl >> 16
mq_nl:
value: bin_mq_nl ^ bin << 16
mapq:
value: mq_nl >> 8
l_read_name:
value: mq_nl ^ (mq << 8)
flag:
value: flag_nc >> 16
no_cigar_op:
value: flag_nc ^ ( flag << 16 )
meta:
id: bgzf
endian: le
seq:
- id: blocks
type: block
repeat: eos
types:
block:
seq:
- id: id1
type: u1
contents: '31'
- id: id2
type: u1
contents: '139'
- id: cm
type: u1
contents: '8'
- id: flg
type: u1
contents: '4'
- id: mtime
type: u4
- id: xfl
type: u1
- id: os
type: u1
- id: xlen
type: u2
- id: extra_subfields
type: extra_subfields
size: xlen
- id: cdata
type: u1
size: extra_subfields.bsize - xlen - 19
process: zlib
- id: crc32
type: u4
- id: isize
type: u4
extra_subfields:
seq:
- id: si1
type: u1
contents: '66'
- id: si2
type: u1
contents: '67'
- id: slen
type: u2
contents: '2'
- id: bsize
type: u2